SMBE satellite meeting on De Novo Gene Birth at Texas A&M this Fall!

It’s happening in two months! I am super excited to co-organize the first meeting on De Novo Gene Birth at Texas A&M. Late registration and abstract submissions will remain open until the end of October, check out the meeting website for more information.
https://agriliferegister.tamu.edu/website/56645/
Thanks to SMBE, NSF, AgriLife Research, the Ecology and Conservation Biology department, the TAMU School of veterinary Medicine & Biomedical Sciences and the Interdisciplinary Graduate Program in Genetics and Genomics at TAMU for their support!
SMBE Italy 2023!

SMBE 2023 in Ferrara was HOT in many ways, certainly for the science.
This was the first SMBE for Adekola, who did a fantastic job on his poster and the presentation.
Welcome to Adekola Owoyemi!

I am very happy to welcome Adekola Owoyemi to the lab as a PhD student in the ECCB program!
Adekola holds both a Master in Biological Sciences and a Graduate Certificate Computational Biology from Auburn University and has joined us this summer. He is currently working on de novo gene discovery, evolution and function in angiosperms.
Yes, it can take me that long to make announcements on this website–To my partial excuse, we have been busy working on a paper that was recently submitted. Adekola has hit the ground running and I can’t wait to celebrate his many future successes!
Convergent evolution paper on C4 chloroplast genes now published in PeerJ

Check out the published version of our paper “Beyond RuBisCO: convergent molecular evolution of multiple chloroplast genes in C4 plants”. https://peerj.com/articles/12791/
Here’ the Abstract:
Background
The recurrent evolution of the C4 photosynthetic pathway in angiosperms represents one of the most extraordinary examples of convergent evolution of a complex trait. Comparative genomic analyses have unveiled some of the molecular changes associated with the C4 pathway. For instance, several key enzymes involved in the transition from C3 to C4 photosynthesis have been found to share convergent amino acid replacements along C4 lineages. However, the extent of convergent replacements potentially associated with the emergence of C4 plants remains to be fully assessed. Here, we conducted an organelle-wide analysis to determine if convergent evolution occurred in multiple chloroplast proteins beside the well-known case of the large RuBisCO subunit encoded by the chloroplast gene rbcL.
Methods
Our study was based on the comparative analysis of 43 C4 and 21 C3 grass species belonging to the PACMAD clade, a focal taxonomic group in many investigations of C4 evolution. We first used protein sequences of 67 orthologous chloroplast genes to build an accurate phylogeny of these species. Then, we inferred amino acid replacements along 13 C4 lineages and 9 C3 lineages using reconstructed protein sequences of their reference branches, corresponding to the branches containing the most recent common ancestors of C4-only clades and C3-only clades. Pairwise comparisons between reference branches allowed us to identify both convergent and non-convergent amino acid replacements between C4:C4, C3:C3 and C3:C4 lineages.
Results
The reconstructed phylogenetic tree of 64 PACMAD grasses was characterized by strong supports in all nodes used for analyses of convergence. We identified 217 convergent replacements and 201 non-convergent replacements in 45/67 chloroplast proteins in both C4 and C3 reference branches. C4:C4 branches showed higher levels of convergent replacements than C3:C3 and C3:C4 branches. Furthermore, we found that more proteins shared unique convergent replacements in C4 lineages, with both RbcL and RpoC1 (the RNA polymerase beta’ subunit 1) showing a significantly higher convergent/non-convergent replacements ratio in C4 branches. Notably, more C4:C4 reference branches showed higher numbers of convergent vs. non-convergent replacements than C3:C3 and C3:C4 branches. Our results suggest that, in the PACMAD clade, C4 grasses experienced higher levels of molecular convergence than C3 species across multiple chloroplast genes. These findings have important implications for our understanding of the evolution of the C4 photosynthesis pathway.
New preprint from the lab
The preprint of our work on convergent evolution of chloroplast proteins in C4 grasses is online on biorXiv.
https://www.biorxiv.org/content/10.1101/2021.08.30.457919v1
I summarized our findings in this thread on Twitter.
New preprint with the amazing @l_jingjia. We tested the hypothesis that convergent evolution of the C4 #photosynthesis left *molecular marks* in multiple #chloroplast #genes. 🧬🌾
The short answer is yes, it did. 🧵#evolution #RuBisCOhttps://t.co/1MJqipuxB2— Claudio Casola (@ccasola) September 1, 2021
Congrats to Shelby Landa for her research proposal approval!
Shout out to Shelby Landa for a great presentation of her master thesis proposal on “Understanding a Major Forest Pest: gene family evolution in wood-boring beetles and annotation of the de novo-assembled Southern Pine Beetle (Dendroctonus frontalis) genome” and passing this hurdle smoothly. On to the thesis and beyond!
New papers out!
Congratulations Jingjia!!
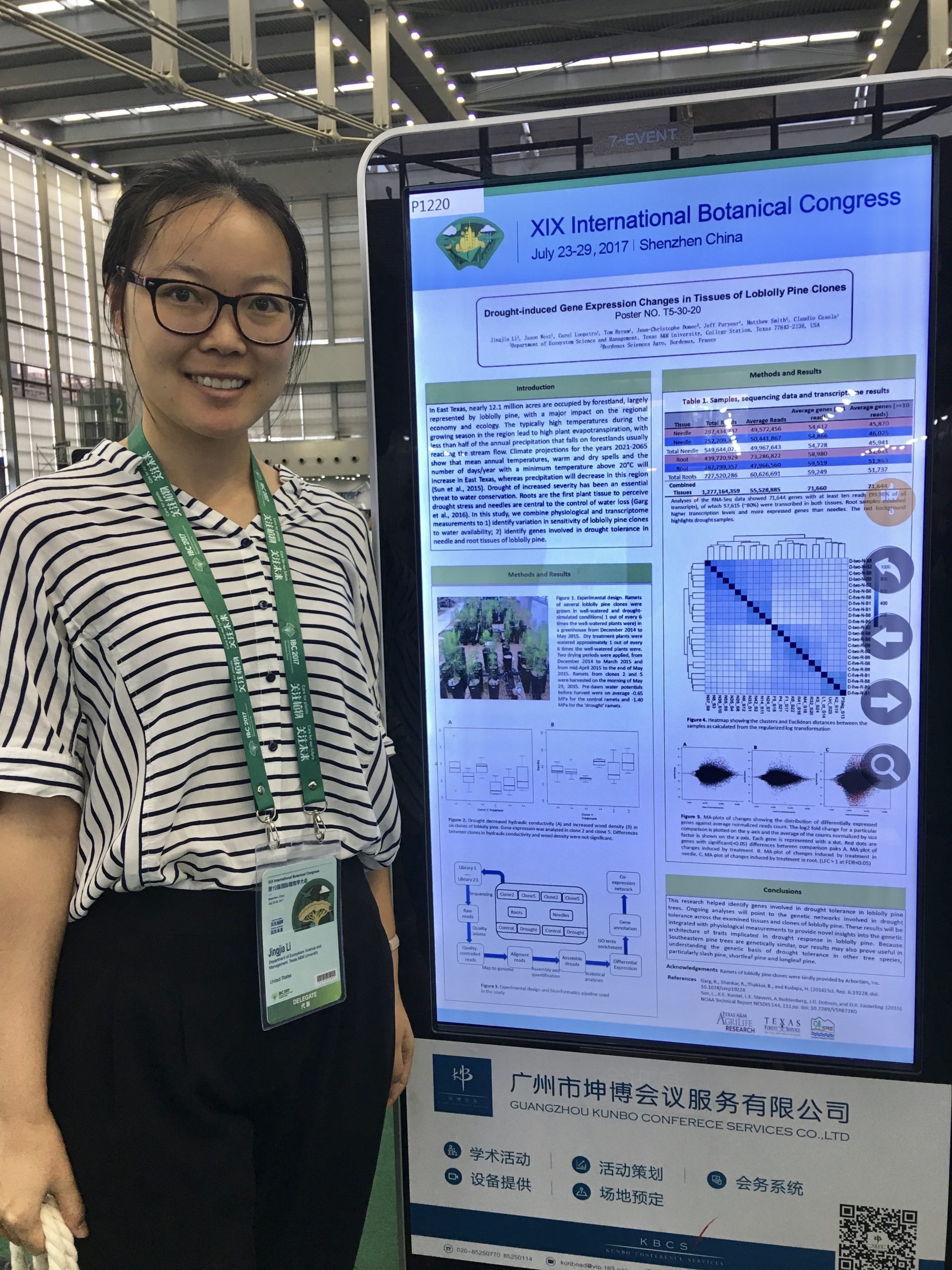 I am very pleased to announce that our Jingjia Li has brilliantly defended her Ph.D. dissertation on the “Functional and evolutionary dynamics of genes involved in drought tolerance in loblolly pine (Pinus taeda L.)”!
I am very pleased to announce that our Jingjia Li has brilliantly defended her Ph.D. dissertation on the “Functional and evolutionary dynamics of genes involved in drought tolerance in loblolly pine (Pinus taeda L.)”!
It has been such a pleasure being the primary advisor of Jingjia in the past four years and to witness her growth as a scientist in my lab. Her work has shed light on many molecular process related to drought tolerance in loblolly. I couldn’t be more excited for her future endevors!
What we stand for
A statement from the Climate, Diversity and Inclusion committee of the Ecology and Conservation Biology Department, of which I am not part of, that I fully endorse.
“We stand with our Black coworkers, students, alumni, and neighbors. We support justice for all lives that have been lost to systemic racism, including those lost to police violence. We acknowledge that systemic racism is present in every sector of our society, and that death and violence are only among the most egregious of the daily manifestations of oppression and prejudice. We see you, we hear you, we believe you, we mourn with you, and we stand with you. Black Lives Matter.”