Our overarching goal is to identify genomic variation between species and populations and to determine their role in adaptation and evolutionary innovation.
We are currently carrying out research in three main areas:
1. New genes, a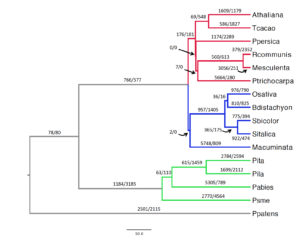 daptation and evolutionary novelties. New genes evolve frequently and play a major role in phenotypic variation, adaptation and the emergence of novel traits. Our primary goals are (1) Determining the contribution of different processes, from gene duplication to de novo gene formation, to the evolution of new genes; and (2) Establishing the biological function of new genes.
daptation and evolutionary novelties. New genes evolve frequently and play a major role in phenotypic variation, adaptation and the emergence of novel traits. Our primary goals are (1) Determining the contribution of different processes, from gene duplication to de novo gene formation, to the evolution of new genes; and (2) Establishing the biological function of new genes.

2. Molecular basis of convergent evolution. Complex phenotypes may evolve independently along multiple lineages that experience similar selective regimes, resulting in convergent adaptations. We are currently interested in studying genetic changes underpinning convergent evolution in C4 plants and beetles.
 3. Genetic basis of drought resistance in loblolly pine. One of our goals is to better understand the genetic basis of drought resistance in loblolly pine (Pinus taeda L.), the most widely planted forest trees in Southeastern U.S. A multi-lab effort is ongoing to integrate physiological measurements, gene expression information and genotyping data to identify genetic variants associated to drought resistance in loblolly pine.
3. Genetic basis of drought resistance in loblolly pine. One of our goals is to better understand the genetic basis of drought resistance in loblolly pine (Pinus taeda L.), the most widely planted forest trees in Southeastern U.S. A multi-lab effort is ongoing to integrate physiological measurements, gene expression information and genotyping data to identify genetic variants associated to drought resistance in loblolly pine.