Overview
The TIGM ES cell clones utilize gene traps for gene inactivation. In this approach, a promoterless marker/reporter gene (for instance encoding Neo, or βgeo) is introduced into ES cells. Selection for expression of the gene requires transcription from a cellular promoter. Consequently, a mutation in a cellular gene and the activity of the tagged gene can be followed by staining for β-galactosidase activity. Unlike gene targeting by homologous recombination, a single gene trap vector can be used in a high-throughput fashion to mutate thousands of individual genes in mouse ES cells, as well as enable the rapid identification of the mutated genes. Large scale sequencing of ES cell clones was conducted and sequence tags have been generated. This information was recently submitted to GenBank to serve the research community at large. A list of the genes we have trapped can be found in our gene trap database. Detailed description of our technology may be found in
Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc Natl Acad Sci U S A. 2003 Nov
Vector Design
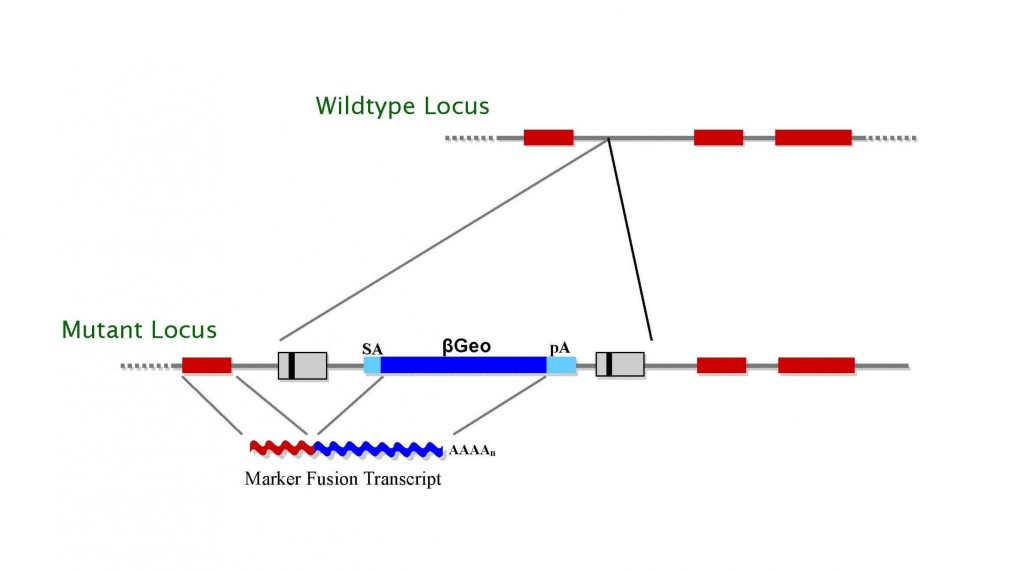
The basic gene trap vectors we have used include a reporter gene downstream of a splice acceptor sequence (Fig.1 and 2). They are designed to function when inserted in an intron, to produce incorrect splicing of the target gene such that all exons downstream of the insertion site are not expressed. The gene trap cassette is inserted in a retroviral vector. Retroviruses insert as a single copy per locus, with no rearrangement of flanking sequences. They have a preference for insertions at the 5′ end of genes, often upstream of the initiator ATG, and the splice acceptor sequence we use does not appear to be bypassed by the RNA-splicing machinery. As a result, the majority of the mutations generated using our gene trap vectors are predicted to lead to null alleles. Analysis of non-embryonic lethal mouse lines demonstrates that gene-trap insertions within both exons and introns of the gene of interest lead to the disruption of the endogenous mRNA transcript in all cases. Of these, >96% show complete absence of WT message, with the remaining hypomorphic lines showing an average reduction in mRNA levels of 91.6%, as measured by quantitative PCR. These data demonstrate that intragenic insertion efficiently disrupts gene transcription in vivo and can be used to reliably predict mutagenicity before mouse production.
C57BL/6 library
TIGM’s C57BL/6 ES cell clones were generated using several different retroviral gene trap vectors that contain either the 5′ selectable marker β-geo, a functional fusion between the β-galactosidase and neomycin resistance genes, for identification of successful gene trap events; or just the neomycin resistance gene alone. The β-geo marker also allows for in vivo expression studies of the trapped gene through in situ hybridization, immunohistochemistry, or β-galactosidase-based assays that can detect the fusion transcript generated as part of the gene trapping process.
Figure 1. Gene trap vectors used in C57BL/6 library (β-geo version shown)*.
We have used high-throughput gene-trapping with retroviral vectors in mouse C57BL/6 ES cells to generate a library of more than 300,000 mutated ES cell clones. Each clone is frozen in duplicate in liquid nitrogen. Wegenerated a tractable sequence tag from all clones using a third replicate of the ES cell clones that was subjected to an automated iPCR-based direct-sequencing protocol. We refer to such sequence as ISTs, which represent genomic sequence of the mutated genes upstream of the genomic insertion site.
* LTR, long terminal repeat; PGK, phosphoglyceratekinase-1 promoter; SD, splice donor sequence; SA, splice acceptor sequence; Neo, neomycin phosphotransferase gene; β-geo, galactosidase/neomycin phosphotransferase fusion gene; pA, polyadenylation sequence; Btk, first exon of the murine Bruton’s tyrosine kinase gene.
29SvEv library
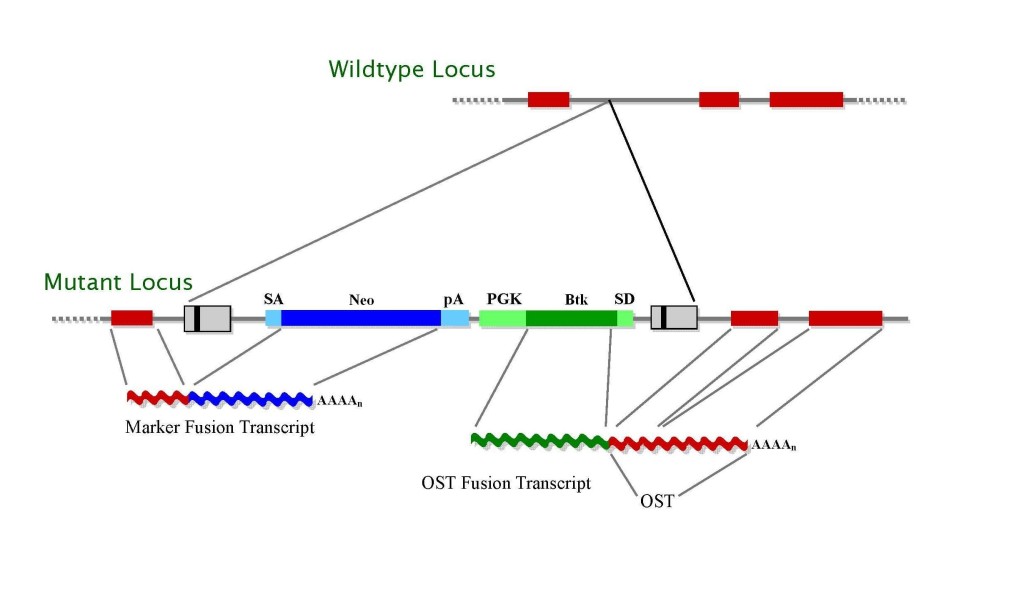
The retroviral vectors (Fig. 2) contain a splice acceptor sequence (SA) followed by a promoterlessselectable marker Neo with a polyadenylationsignal (pA). Insertion of the retroviral vector into an expressed gene leads to the splicing of the endogenous upstream exonsinto this cassette to generate a fusion transcript. The vectors also contain a promoter that is active in ES cells [such as that of the mouse phosphoglyceratekinase (
Pgk) gene] followed by a first exon (such as that of the Bruton’s Tyrosine Kinase (
Btk) gene) upstream of a splice donor (SD) signal. Splicing from this signal to the exonsdownstream of the insertion gives rise to a fusion transcript that can be used to generate a sequence tag (OST) of the trapped gene by 3′ RACE (
Zambrowicz, 1998). The
Btk exon contains termination codons in all reading frames to prevent translation of downstream fusion transcripts.
Figure 2. Gene trap vectors used in 129SvEv library*
High-throughput gene-trapping with retroviral vectors has been used in mouse 129SvEv ES cells to generate a library of 522,666 mutated ES cell clones. Each clone is frozen in duplicate in liquid nitrogen. Tractable sequence tags have been generated from 271,860 clones (52%), using a third replicate of the ES cell clones that was subjected to an automated RT-PCR-based direct-sequencing protocol (
Zambrowicz, 2003). Such sequences are being referred to as OSTs, which represent cDNAsequence of the mutated genes downstream of the genomic insertion site (Fig. 2).
* LTR, long terminal repeat; PGK, phosphoglyceratekinase-1 promoter; SD, splice donor sequence; SA, splice acceptor sequence; Neo, neomycin phosphotransferase gene; β-geo, galactosidase/neomycin phosphotransferase fusion gene; pA, polyadenylation sequence; Btk, first exon of the murine Bruton’s tyrosine kinase gene.
Targeted mutations
In addition to our gene trap collections, TIGM can provide more than 2000 mutant mouse lines in 129/SvEv x C57BL/6 background produced by homologous recombination-based gene targeting.
The targeting vectors (Fig. 3) contain Internal Ribosome Entry Site (IRES) followed by a promoterless selectable marker β-geo, a functional fusion between the β-galactosidase and neomycin resistance genes, for identification of successful homologous recombination events. The β-geo marker also allows for in vivo expression studies of the targeted gene through in situ hybridization, immunohistochemistry, or β-galactosidase-based assays that can detect the fusion transcript generated as part of the gene targeting process. The IRES/βGEo/PloyA cassette is positioned to take advantage of the target gene’s exon’s natural splice donor and create a fusion transcript.
Figure 3. Gene targeting*

* IRES, Internal Ribosome Entry Site; β-geo, galactosidase/neomycin phosphotransferase fusion gene; pA, polyadenylation sequence;



